Grid storage¶
In this page we will talk about the Grid storage facilities, the tools to interact with it and the method to handle data that is stored on tape.
Contents
About Grid storage¶
Each cluster on the Grid is equipped with a Storage Element or SE where data is stored. The Grid storage is useful for applications that handle large amount of data that can not be sent with the job Sandbox or stored in a pilot job database.
You can interact with the Grid storage from the UI or from a Worker Node, within your running job. The scripts that can access the Grid storage can be submitted from:
To use the Grid storage you must:
- Have a personal Grid certificate [1]_
- Be member of a VO for which we have allocated storage space.
You can access the Grid storage with Grid Storage clients, through interfaces that speak protocols like SRM, GridFTP, GSIdCap or WebDAV. With these storage clients you can:
- list directories and files
- read (download) files
- write (upload) files
- delete files or directories
- stage files (copy them from tape to disk for faster reading)
| [1] | It is technically possible to access the dCache Grid storage without certificate, by using WebDAV with username/password authentication. We don’t recommend this: authentication with username/password is less secure, and WebDAV is slower than GridFTP. |
Storage types¶
There are two storage types available on the Dutch Grid sites:
- The dCache storage element located at SURFsara and accessible from any Grid site.
- The DPM storage elements located at each LSG cluster and accessible only by the Life Science Grid users.
dCache¶
The storage element located at SURFsara is accessible from any Grid cluster or UI. It uses the dCache system for storing and retrieving huge amounts of data, distributed among a large number of server nodes. It consists of magnetic tape storage and hard disk storage and both are addressed by a common file system. See dCache specifications for details about our dCache instance.
DPM¶
The storage elements located at the various Life Science Grid clusters are accessible only by the LSG users. The LSG clusters have local storage that uses DPM (short for Disk Pool Manager).
Note
The DPM storage is only disk storage and does not support tape back-end. In opposite, the dCache central storage has both disk and tape.
Grid file identifiers¶
You can refer to your files on the Grid with different ways depending on which of the available Storage clients you use to manage your files:
Transport URL or TURL¶
Examples:
# lsgrid user homer stores the file zap.tar on dCache storage
gsiftp://gridftp.grid.sara.nl:2811/pnfs/grid.sara.nl/data/lsgrid/homer/zap.tar
# same, but with a WebDAV TURL
https://webdav.grid.sara.nl/pnfs/grid.sara.nl/data/lsgrid/homer/zap.tar
# lsgrid user homer stores the file zap.tar on DPM storage at lumc cluster
gsiftp://gb-se-lumc.lumc.nl:2811/dpm/lumc.nl/home/lsgrid/homer/zap.tar
Clients for TURLs
- uberftp
- globus
- gfal
- fts
- globusonline
Storage URL or SURL¶
Examples:
# lsgrid user homer stores the file zap.tar on dCache storage
srm://srm.grid.sara.nl:8443/pnfs/grid.sara.nl/data/lsgrid/homer/zap.tar
# lsgrid user homer stores the file zap.tar on DPM storage at lumc cluster
srm://gb-se-lumc.lumc.nl:8446/dpm/lumc.nl/home/lsgrid/homer/zap.tar
Clients for SURLs
- srm
- gfal
- fts
- lcg-lfn-lfc
Logical File Name (LFN) and Grid Unique Identifier (GUID)¶
These identifiers correspond to logical filename such as lfn:/grid/lsgrid/homer/zap.tar
Note
The SURLs and TURLs contain information about where a physical file is located. In contrast, the GUIDs and LFNs identify a logical filename irrespective of its location. You only need to use these if you work with Data replication on multiple LSG sites.
Default ports¶
dCache¶
| Protocol | Host(s) and port(s) | Remark |
|---|---|---|
| SRM | srm://srm.grid.sara.nl:8443 | |
| GridFTP | gsiftp://gridftp.grid.sara.nl:2811 | Data channel port range: 20000-25000 |
| WebDAV | https://webdav.grid.sara.nl:443 | See webdav client for details |
| https://webdav.grid.sara.nl:2880 | ||
| https://webdav.grid.sara.nl:2881 | ||
| GSIdCap | gsidcap://gsidcap.grid.sara.nl:22128 | |
| xroot | xrootd.grid.sara.nl:1094 | Used by CERN only |
DPM¶
| Protocol | Host(s) and port(s) (examples) | Remark |
|---|---|---|
| SRM | srm://gb-se-lumc.lumc.nl:8446 | |
| GridFTP | gsiftp://gb-se-lumc.lumc.nl:2811 | Data channel port range: 20000-25000 |
For an overview of all life science clusters and their DPM storage elements, see Cluster details
Storage clients¶
The InputSandbox and OutputSandbox attributes in the JDL file are the basic way to move files to and from the User Interface (UI) and the Worker Node (WN). However, when you have large files (from about 100 MB and larger) then you should not use these sandboxes to move data around. Instead you should use the Storage types and work with several Storage clients.
In this section we will show the common commands to use the various storage clients.
Note
From the many Grid storage clients, we recommend you to use the uberftp client, globus client or gfal client. These tools have a clean interface, and their speed is much better on our systems compared with their srm-* equivalents.
| protocols | |||||||
|---|---|---|---|---|---|---|---|
| Client | SRM | GridFTP | GSIdCap | WebDAV | 3rd party | Speed | Tape control [1]_ |
| uberftp client | – | yes | – | – | – | high | – |
| globus client | – | yes | – | – | – | high | – |
| srm client | yes | [2] | [2] | [2] | – | yes | |
| gfal client | yes | yes | – | – | – | yes | |
| webdav client | – | – | – | yes | – | – | |
| fts client | yes | yes | – | yes | yes | high | yes |
| globusonline client | – | yes | – | – | yes | high | – |
| lcg-lfn-lfc clients (not recommended) | yes | [2] | – | – | – | – | |
| [1] | Examples of tape control: staging a file from tape to disk, or get it locality (tape or disk). |
| [2] | (1, 2, 3, 4) SRM and LCG commands use the SRM protocol for metadata level operations and switch to another protocol like GridFTP for file transfers. This may cause protocol overhead. For example, authentication needs to be done twice: once for each protocol. |
Staging files¶
The dCache storage at SURFsara consists of magnetic tape storage and hard disk storage. If your quota allocation includes tape storage, then the data stored on magnetic tape has to be copied to a hard drive before it can be used. This action is called Staging files or ‘bringing a file online’.
| Locality | Meaning |
|---|---|
| ONLINE | The file is only on disk |
| NEARLINE | The file is only on tape; it should be staged before reading it |
| ONLINE_AND_NEARLINE | The file is both on disk and on tape |
Staging a single file¶
Note
For all staging operations you need to have a valid proxy, see StartGridSession.
Here is an example of how to stage a single file:
$srm-bring-online srm://srm.grid.sara.nl/pnfs/grid.sara.nl/data/lsgrid/test
srm://srm.grid.sara.nl/pnfs/grid.sara.nl/data/lsgrid/test brought online, use request id 424966221 to release
Don’t use this method to stage multiple files. Use the stage.py example below instead, because it is much more efficient.
How to display the locality:
$srmls -l srm://srm.grid.sara.nl/pnfs/grid.sara.nl/data/lsgrid/test | grep locality
locality:ONLINE_AND_NEARLINE
Staging groups of files¶
The example below shows how to stage a list of files with known SURLs.
Copy and untar the tarball
staging scriptsto your UI home directory.Create a proxy on the UI:
$startGridSession lsgridThe file paths should be listed in a file called
fileswith the following format:/pnfs/grid.sara.nl/data/...Let’s say that you have a list of SURLs that you want to stage. Convert the list of SURLs in the
datasets/example.txtfile to the desired/pnfsformat:$grep --only-matching '/pnfs/grid.sara.nl.*' datasets/example.txt > files
Display the locality of the files with:
$python state.pyStage the files:
$python stage.py
This script stages a number of files from tape. You can change the pin lifetime in the stage.py script by changing the srmv2_desiredpintime attribute in seconds.
Monitor staging activity¶
Once you submit your stage requests, you can use the gfal scripts to monitor the status or check the webpage below that lists all the current staging requests:
Unpin a file¶
Your files may remain online as long as there is free space on the disk pools. When a pool group is full and free space is needed, dCache will purge the least recently used cached files. The tape replica will remain on tape.
The disk pool where your files are staged has limited capacity and is only meant for data that a user wants to process on a Grid site. When you pin a file you set a pin lifetime. The file will not be purged until the pin lifetime has expired. Then the data may be purged from disk, as soon as the space is required for new stage requests. When the disk copy has been purged, it has to be staged again in order to be processed on a Worker Node.
When a pool group is full with pinned files, staging is paused. Stage requests will just wait until pin lifetimes for other files expire. dCache will then use the released space to stage more files until the pool group is full again. When this takes too long, stage requests will time out. So pinning should be used moderately.
When you are done with your processing, we recommend you release (or unpin) all the files that you don’t need any more. In order to unpin a file, run from the UI:
$srm-release-files srm://srm.grid.sara.nl:8443/pnfs/grid.sara.nl/data/lsgrid/homer/zap.tar # replace with your SURL
This command will initiate unpinning of file zap.tar (even if you submitted multiple pin requests) and the file will remain cached but purgeable until new requests will claim the available space. It is an optional action, but helps a lot with the effective system usage.
Warning
At the moment neither the srm-bring-online nor the python gfal scripts can effectively release a file if there are multiple pin requests. Please use srm-release-files.
SRM interaction example diagram¶
Here is a sequence diagram that illustrates how the SRM commands interact with the Grid storage.
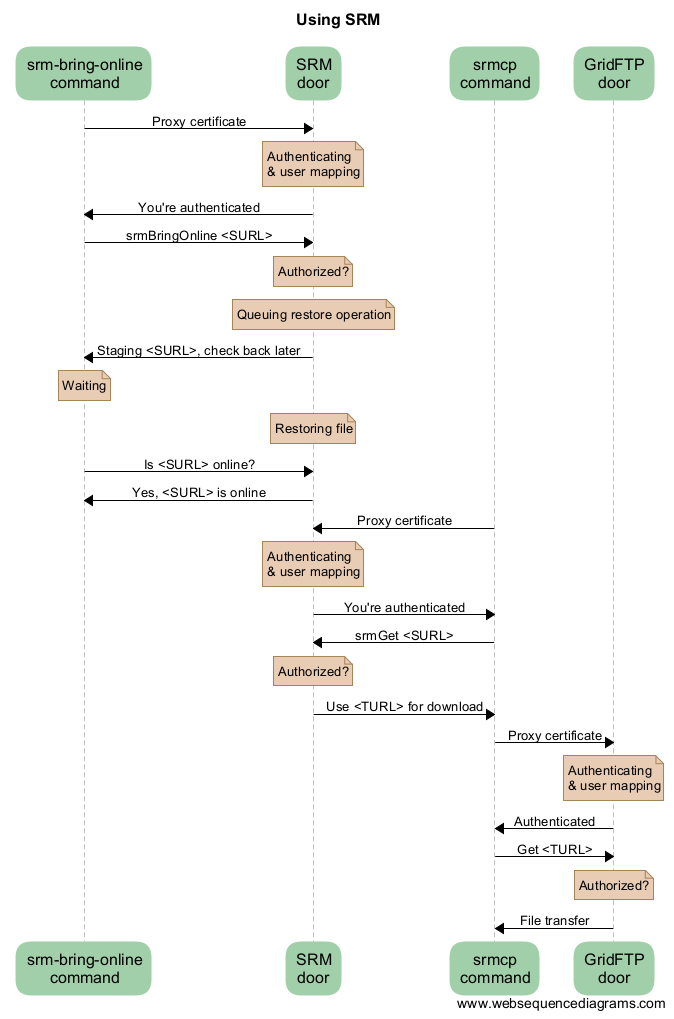
As you can see from this diagram, there can be a lot of overhead per file. For this reason, Grid storage performs best with large files. We recommend to store files of several megabytes to gigabytes. Some users have files of more than 2 terabytes, but that may be impractical on other systems with limited space like worker nodes.
Importing large amounts of data¶
The Data Ingest Service is a SURFsara service for researchers who want to store or analyze large amounts of data at SURFsara. The service is convenient for users who lack sufficient bandwidth or who have stored their data on a number of external hard disks.

